Scientists have found a way to trap and break down “forever chemicals” instead of just moving them elsewhere.



Psychedelic research is entering a new era, offering hope for chronic pain, anxiety, hearing loss, and more.
Check out the latest findings here.
New studies show that psychedelics could unlock safer treatments for chronic pain, mood disorders, and even neuroprotection. Explore the latest breakthroughs.

Researchers at LMU have uncovered how ribosomes, the cell’s protein builders, also act as early warning sensors when something goes wrong inside a cell.
When protein production is disrupted, and ribosomes begin to collide, a molecule called ZAK detects the pileup and switches on protective stress responses.
Ribosomes as protein builders and stress sensors.

A collaborative team has achieved the first monolithic 3D chip built in a U.S. foundry, delivering the densest 3D chip wiring and order-of-magnitude speed gains.
Engineers at Stanford University, Carnegie Mellon University, University of Pennsylvania, and the Massachusetts Institute of Technology have collaborated with SkyWater Technology, the largest exclusively U.S.-based pure-play semiconductor foundry, to develop a novel multilayer computer chip whose architecture could help usher in a new era of AI hardware and domestic semiconductor innovation.
Unlike today’s largely flat, 2D chips, the new prototype’s key ultra-thin components rise like stories in a tall building, with vertical wiring acting like numerous high-speed elevators that enable fast, massive data movement. Its record-setting density of vertical connections and carefully interwoven mix of memory and computing units help the chip bypass the bottlenecks that have long slowed improvement in flat designs. In hardware tests and simulations, the new 3D chip outperforms 2D chips by roughly an order of magnitude.
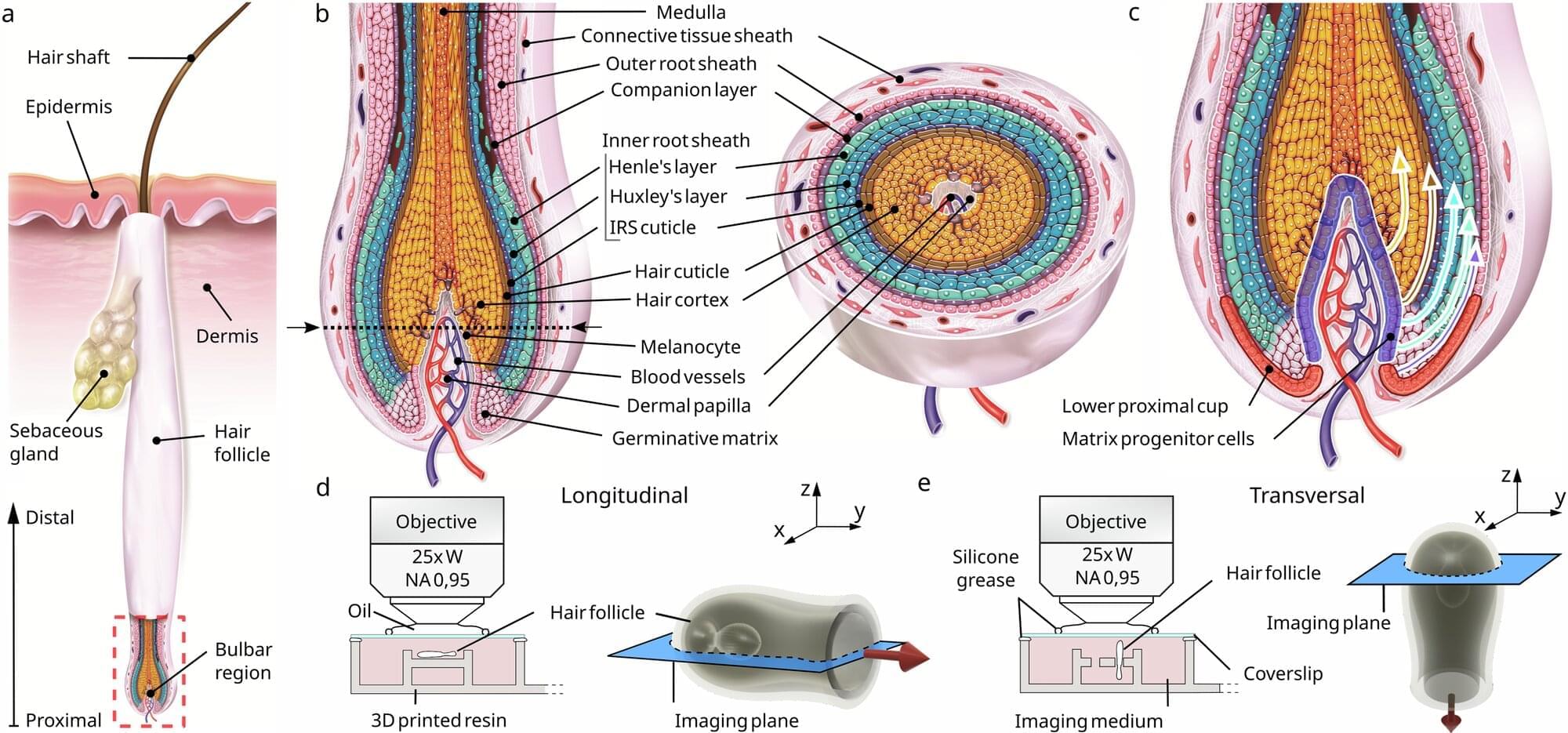
Scientists have found that human hair growth does not grow by being pushed out of the root; it’s actually pulled upward by a force associated with a hidden network of moving cells. The findings challenge decades of textbook biology and could reshape how researchers think about hair loss and regeneration.
The team, from L’Oréal Research & Innovation and Queen Mary University of London, used advanced 3D live imaging to track individual cells within living human hair follicles kept alive in culture. The study, published in Nature Communications, shows that cells in the outer root sheath—a layer encasing the hair shaft—move in a spiral downward path within the same region from where the upward pulling force originates.
Dr. Inês Sequeira, Reader in Oral and Skin Biology at Queen Mary and one of the lead authors, said, “Our results reveal a fascinating choreography inside the hair follicle. For decades, it was assumed that hair was pushed out by the dividing cells in the hair bulb. We found that instead that it’s actively being pulled upwards by surrounding tissue acting almost like a tiny motor.”
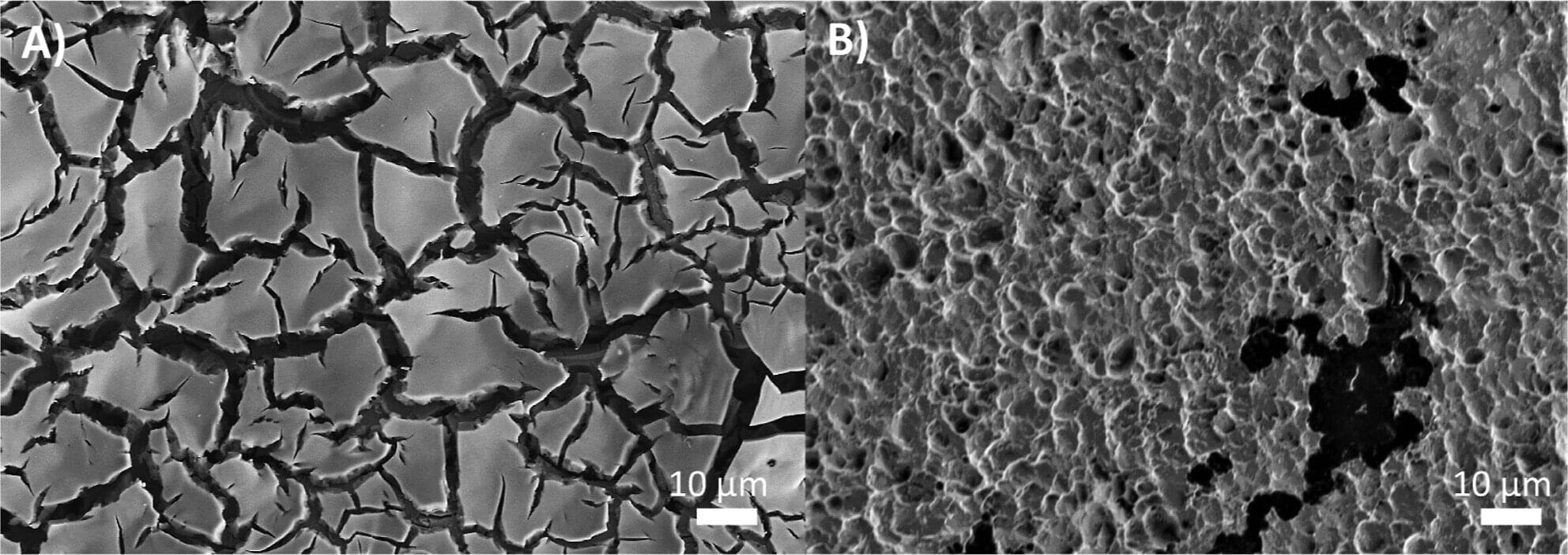
University of Pittsburgh researchers have made an important step toward providing hospitals and water treatment facilities with a safer, greener alternative to chlorine-based disinfection.
The team, which includes scientists from Drexel University and Brookhaven National Laboratory, uncovered key design principles for catalysts that can generate ozone, a disinfecting agent, on demand. The research is published in the journal ACS Catalysis.
This breakthrough addresses a critical challenge in water sanitation. Chlorine, commonly used to kill bacteria on surfaces and in water—including most municipal drinking water—is hazardous to transport and store, and its byproducts can be carcinogenic. These risks limit its use and motivate the search for safer disinfectants.
“With GB200 NVL72 and Together AI’s custom optimizations, we are exceeding customer expectations for large-scale inference workloads for MoE models like DeepSeek-V3,” said Vipul Ved Prakash, cofounder and CEO of Together AI. “The performance gains come from NVIDIA’s full-stack optimizations coupled with Together AI Inference breakthroughs across kernels, runtime engine and speculative decoding.”
This performance advantage is evident across other frontier models.
Kimi K2 Thinking, the most intelligent open-source model, serves as another proof point, achieving 10x better generational performance when deployed on GB200 NVL72.
Nuclear power company TerraPower has passed the Nuclear Regulatory Commission staff’s final safety evaluation for a permit to build a reactor in Wyoming. The Washington-based company backed by Bill Gates and NVIDIA could be the first to deploy a utility-scale, next-generation reactor in America.
TerraPower’s Natrium design pairs a small modular reactor (SMR) with an integrated thermal battery. The SMR generates 345 megawatts of continuous electrical power. The thermal battery, which stores excess heat in molten salt, allows the system to surge its output to 500 megawatts for more than five hours, generating enough energy to power 400,000 homes at maximum capacity.
“Today is a momentous occasion for TerraPower, our project partners and the Natrium design,” said company CEO Chris Levesque in a statement issued Monday. The favorable assessment “reflects years of rigorous evaluation, thoughtful collaboration with the NRC, and an unwavering commitment to both safety and innovation.”